The greatest map?
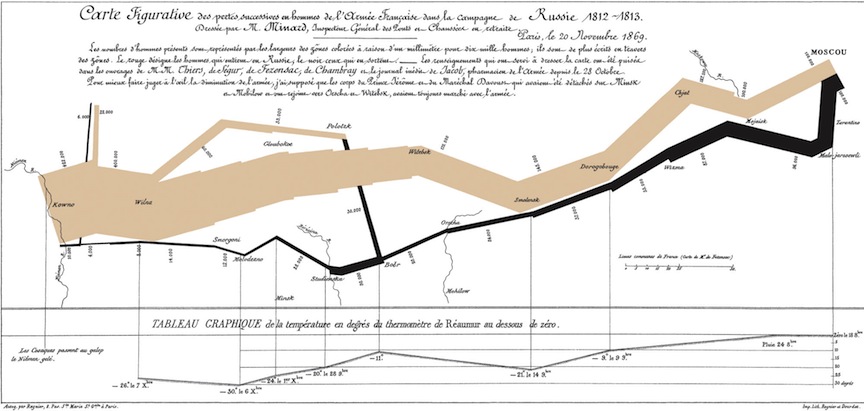
Minard’s Map of Napolean’s Russian Campaign of 1812
- Edward Tufte might say yes
- enormous amount of data laid out in organized, understandable fashion
- multi-media presentations of lines, numbers, pictures, narrative
- no proprietary platform, easily accessible by multiple users
- entirely content, e.g. no drop shadows, bullet indents etc…
- genuinely interactive (people instinctively look for familiar areas, etc…)